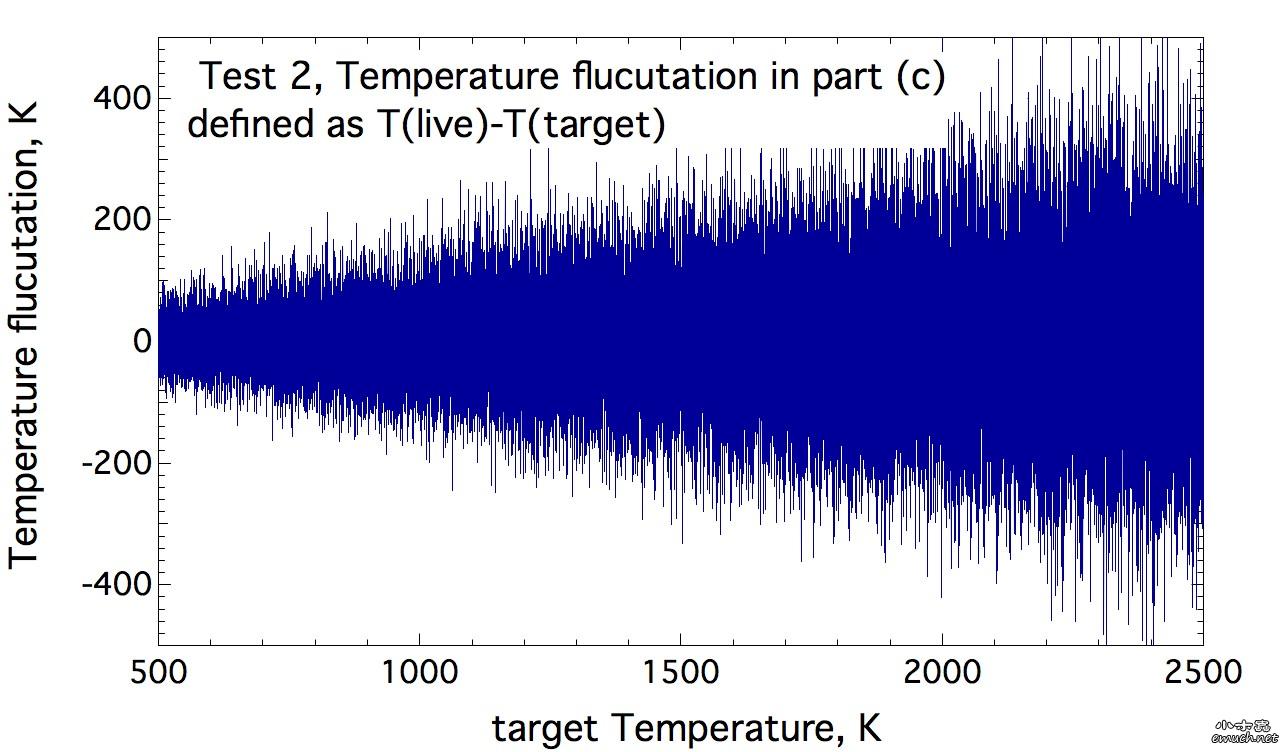
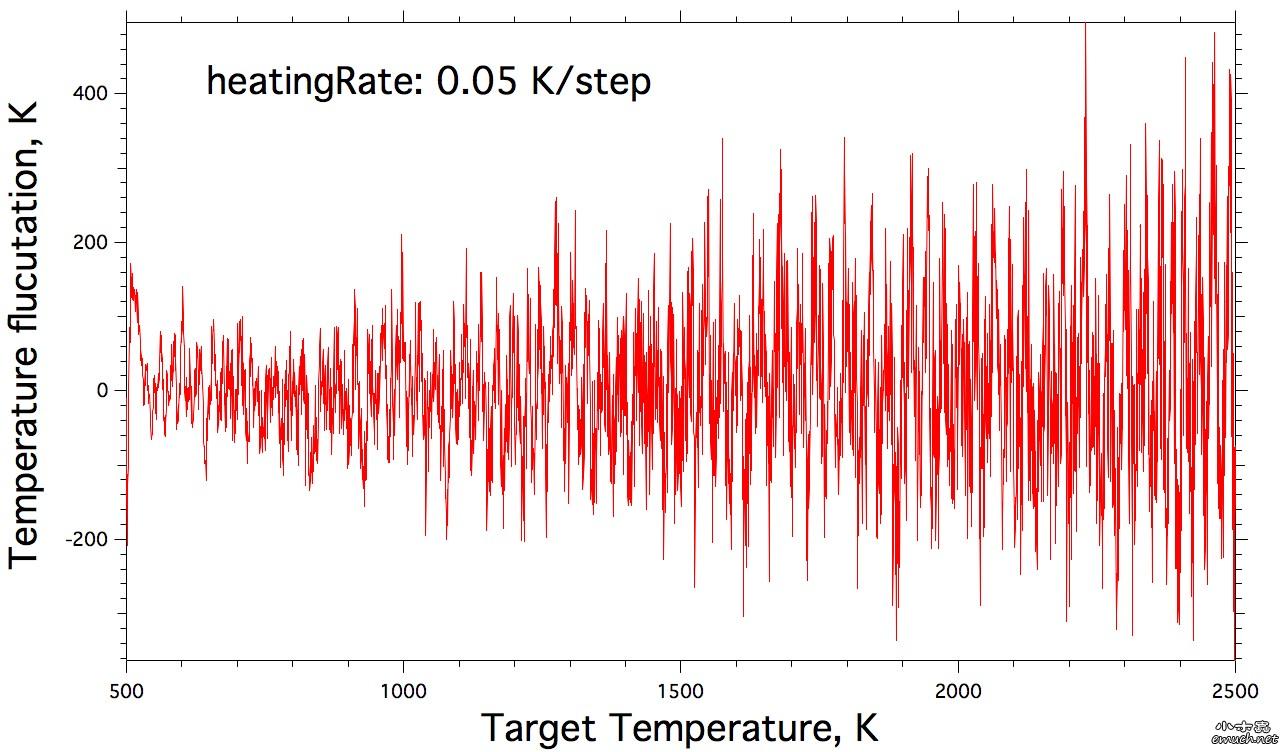
| 查看: 6023 | 回复: 15 | ||||||||||||
| 本帖产生 2 个 模拟EPI ,点击这里进行查看 | ||||||||||||
qphll金虫 (正式写手)
|
[交流]
【分享】尝试lammps中, 分享中... 已有9人参与
|
|||||||||||
|
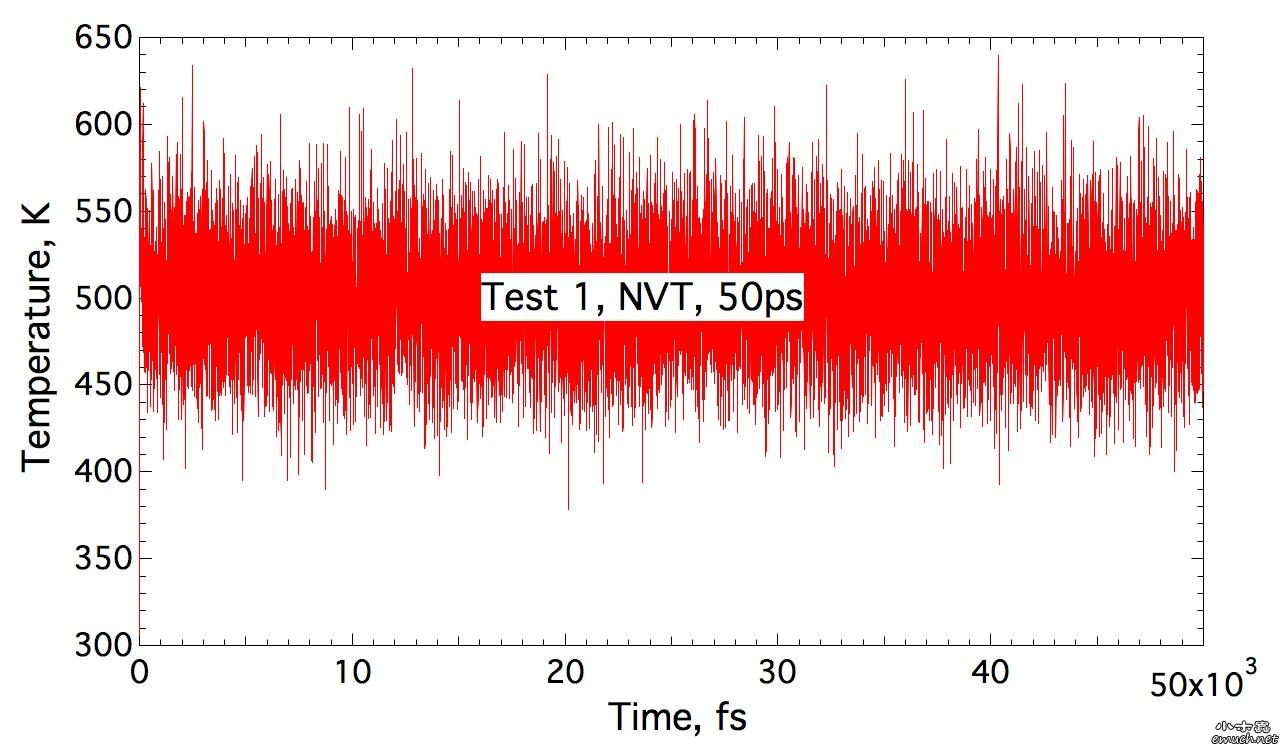
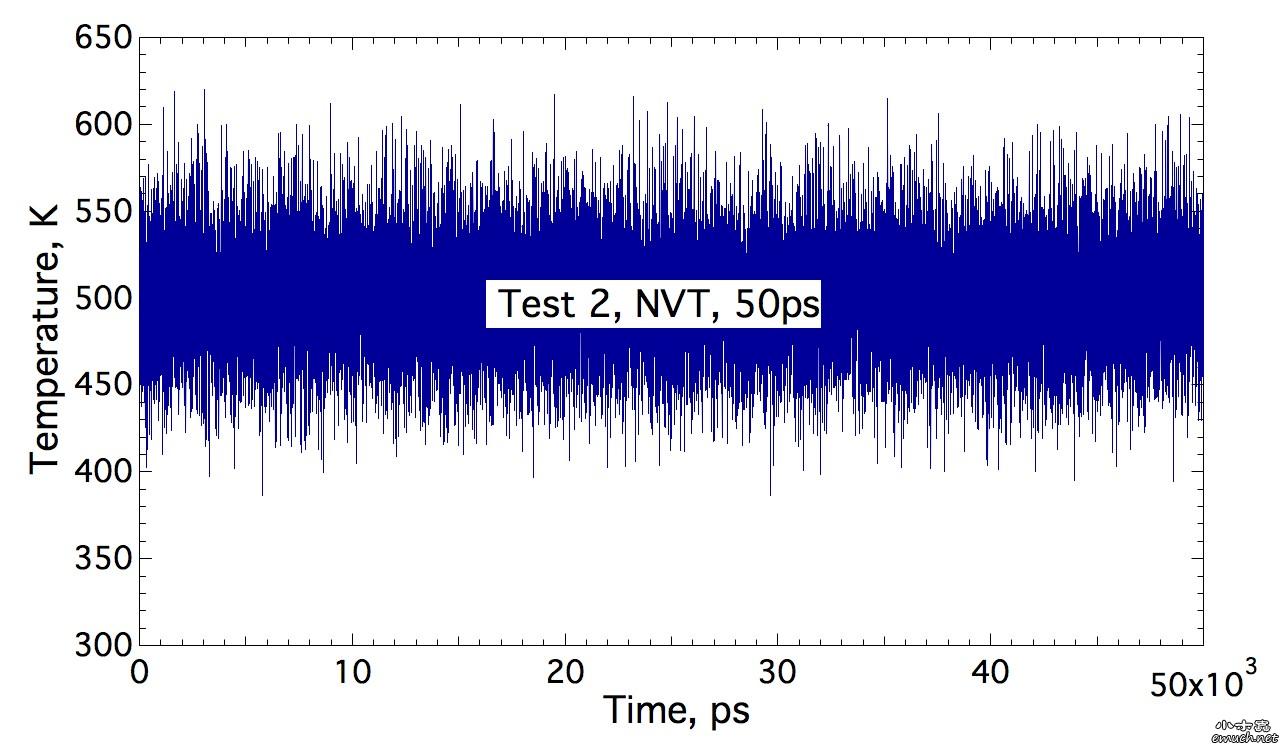
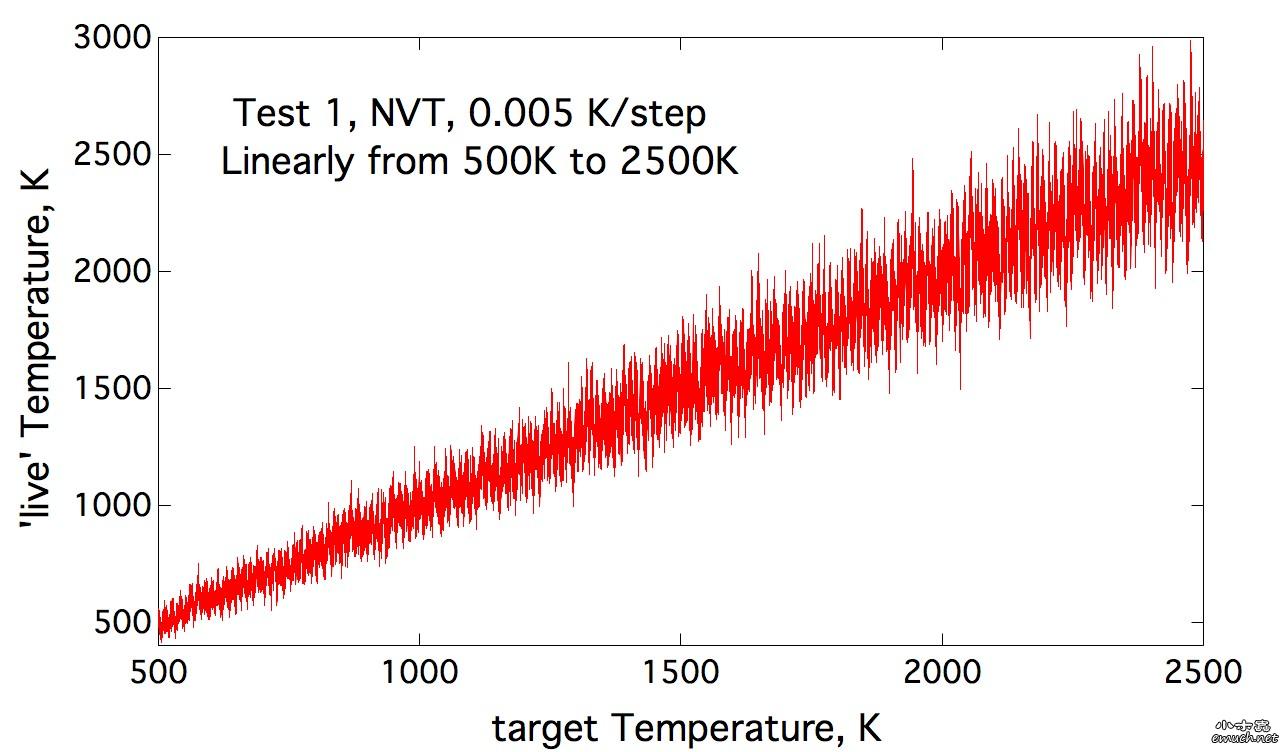
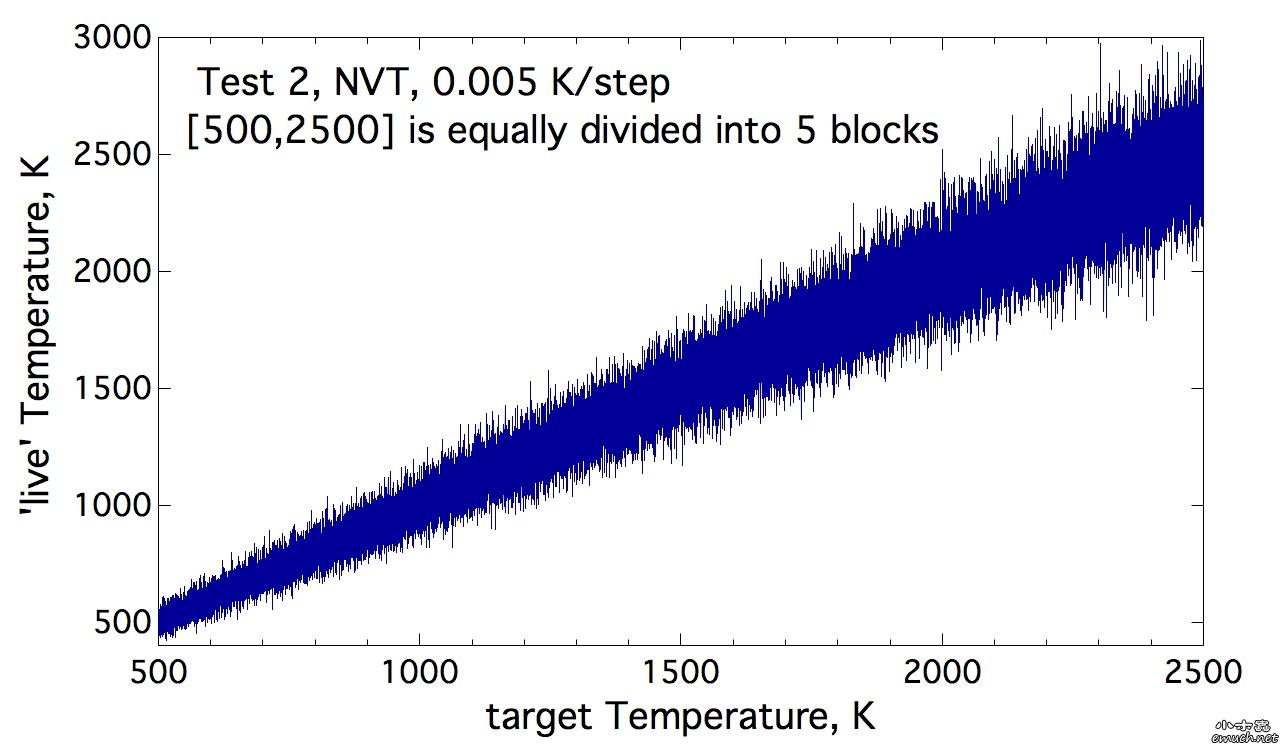
Lesson 1 Loop inside Lammps input script 想要尝试在lammps的input script里面做循环, 结果因为一个小问题, 捣鼓了我几个小时, 这才完全通过测试. 分享一下. (1) input script里面的循环块 include tempfile include tempfile2 variable i loop 10 label loopa fix 2 all nvt temp ${mytemp} ${mytemp2} 100.0 run 200 unfix 2 next mytemp next mytemp2 next i jump SELF loopa 这个的SELF是让程序执行到这里, 跳回自己, 然后从标签 loopa开始执行. 当然loopa是随便取的, 你可以用CHN来做label. 另外, 在某些情况下, 如果要让c++ rewind, 那么最好在执行脚本里面这样写: lmp -in script 而不是 lmp < script 否则, 你人品不好的时候, 会出问题, 哈哈. (2) tempfile 和 tempfile2是在同目录下的另外两个文件. tempfile的文件内容是: variable mytemp index 500.0 700.0 900.0 1100.0 1300.0 1500.0 1700.0 1900.0 2100.0 2300.0 注意, 只有一行! 至于lammps能读多长的一行, 我还没有测试. tempfile2的文件内容, 也是一行: variable mytemp2 index 700.0 900.0 1100.0 1300.0 1500.0 1700.0 1900.0 2100.0 2300.0 2500.0 折腾我的问题是, 我原先在这两个tempfile文件中, 数值之间用逗号分隔, 但是事实上, 是需要用空格分隔的. (3) 如果你需要做的循环不是很多, 那么不需要额外准备tempfile 和 tempfile2文件. 而只是需要在 input script中这样做: variable mytemp index 500.0 700.0 900.0 1100.0 1300.0 1500.0 1700.0 1900.0 2100.0 2300.0 #variable mytemp2 index 700.0 900.0 1100.0 1300.0 1500.0 1700.0 1900.0 2100.0 2300.0 2500.0 variable i loop 10 label loopa fix 2 all nvt temp ${mytemp} ${mytemp2} 100.0 run 200 unfix 2 next mytemp next mytemp2 next i jump SELF loopa 当然, 这里不需要原先的这两句 include语句了. 总结一下你需要熟悉的命令: variable, include, jume, next End of Lesson 1. ENJOY. [ Last edited by qphll on 2010-12-3 at 11:54 ] |
» 收录本帖的淘帖专辑推荐
 资源收集 资源收集 |  材料计算模拟实用技巧 材料计算模拟实用技巧 |  分子模拟 分子模拟 |  MD分子动力学 MD分子动力学 |
 我学习计算的一些帖子 我学习计算的一些帖子 |  分子动力学 分子动力学 |  量化 量化 |  LAMMPS LAMMPS |
 关于Lammps 关于Lammps |
» 本帖已获得的红花(最新10朵)
» 猜你喜欢
 为什么中国大学工科教授们水了那么多所谓的顶会顶刊,但还是做不出宇树机器人?
已经有9人回复
为什么中国大学工科教授们水了那么多所谓的顶会顶刊,但还是做不出宇树机器人?
已经有9人回复
 版面费该交吗
已经有9人回复
版面费该交吗
已经有9人回复
 体制内长辈说体制内绝大部分一辈子在底层,如同你们一样大部分普通教师忙且收入低
已经有13人回复
体制内长辈说体制内绝大部分一辈子在底层,如同你们一样大部分普通教师忙且收入低
已经有13人回复
 面上可以超过30页吧?
已经有4人回复
面上可以超过30页吧?
已经有4人回复
 “人文社科而论,许多学术研究还没有达到民国时期的水平”
已经有5人回复
“人文社科而论,许多学术研究还没有达到民国时期的水平”
已经有5人回复
 什么是人一生最重要的?
已经有4人回复
什么是人一生最重要的?
已经有4人回复
» 本主题相关价值贴推荐,对您同样有帮助:
 在Lammps中安装xmovie工具时,Makefile文件怎么修改X库路径
已经有3人回复
在Lammps中安装xmovie工具时,Makefile文件怎么修改X库路径
已经有3人回复
 lammps中固定键长
已经有4人回复
lammps中固定键长
已经有4人回复
 【求助】lammps安装过程中,在安装fftw的过程中遇到问题
已经有8人回复
【求助】lammps安装过程中,在安装fftw的过程中遇到问题
已经有8人回复
 【讨论】想学lammps是否应该先学习C++
已经有28人回复
【讨论】想学lammps是否应该先学习C++
已经有28人回复
 【讨论】lammps-怪异的group温度
已经有9人回复
【讨论】lammps-怪异的group温度
已经有9人回复
 【讨论】lammps里的boundary的位置能够随意更改?
已经有14人回复
【讨论】lammps里的boundary的位置能够随意更改?
已经有14人回复
 【讨论】lammps中利用velocity命令无法控制住速度问题
已经有11人回复
【讨论】lammps中利用velocity命令无法控制住速度问题
已经有11人回复
 【求助】lammps中多元化合物的晶格创建问题
已经有14人回复
【求助】lammps中多元化合物的晶格创建问题
已经有14人回复

qphll
金虫 (正式写手)
- 模拟EPI: 10
- 应助: 18 (小学生)
- 金币: 2708.7
- 散金: 2294
- 红花: 8
- 帖子: 554
- 在线: 227.1小时
- 虫号: 89654
- 注册: 2005-08-29
- 性别: GG
- 专业: 碳素材料与超硬材料
★ ★ ★ ★ ★ ★ ★ ★ ★ ★
ghcacj(金币+10):谢谢 2010-12-10 10:41:06
ghcacj(金币+10):谢谢 2010-12-10 10:41:06
|
Q: 如何在跑完reax MD以后, 判断是否有反应发生了呢? A: (1) 在input script文件中使用命令 'fix reax/bonds' 输出成键信息, 具体请查询手册; (2) 使用这里给出的小程序来读取output文件, 将提取出来的信息作图判断. 请根据自己的体系, 自行修改给出的小程序. 我还没来得及改进.... !# DEC.9, 2010 !# QPHLL !# !# This is a program to read the output from 'fix reax/bond', TPRD, Lammps !# The output is saved into file "bonds.connect", where each image is divided !# into three parts: !# !# (1) Head, 7 Lines; !# (2) Body, No._of_atom Lines; !# (3) Tail, 1 Line !# !# The total number of images is related with the output frequence and number of iterations. !# In this case, it is "number of iteration+1". !# !# Each line in Body part is made up of the following parameters: !# id, type, nb, id_1, id_2, ... id_nb, mol, bo_1, bo_2, ... bo_nb, abo, nlp, q !# abo = atomic bond order !# nlp = number of lone pairs !# q = atomic charge !# !# PLEASE DOUBLE CHECK YOUR OWN LAMMPS INPUT SCRIPT & OUTPUT AND MAKE CORRESPONDING CHSNGES program main implicit none integer I, J, K, L integer image, natom integer headline, tailline integer id, atype, nb, bd1, bd2, bd3, mol double precision bo1, bo2, bo3, abo, nlp, q open (unit=10, file='bonds.connect') open (unit=20, file='N129.txt', status='unknown') open (unit=21, file='N133.txt', status='unknown') open (unit=22, file='N137.txt', status='unknown') open (unit=23, file='N141.txt', status='unknown') open (unit=24, file='N145.txt', status='unknown') open (unit=25, file='N149.txt', status='unknown') open (unit=26, file='N153.txt', status='unknown') open (unit=27, file='N157.txt', status='unknown') image = 2000 headline = 7 tailline = 1 natom = 160 do I = 1, image+1 do J = 1, headline read(10,*) end do do K = 1, natom read(10,*) id, atype, nb write(*,*) id, atype, nb if (atype .eq. 4) then backspace 10 read(10,*) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q write(*,*) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q if (id .eq. 129) then write(20, 200) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 133) then write(21, 200) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 137) then write(22, 200) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 141) then write(23, 200) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 145) then write(24, 200) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 149) then write(25, 200) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 153) then write(26, 200) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 157) then write(27, 200) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q 200 format(7I4, 6f14.3) endif endif enddo do L =1,tailline read(10,*) enddo enddo end program main |

5楼2010-12-10 01:47:12
qphll
金虫 (正式写手)
- 模拟EPI: 10
- 应助: 18 (小学生)
- 金币: 2708.7
- 散金: 2294
- 红花: 8
- 帖子: 554
- 在线: 227.1小时
- 虫号: 89654
- 注册: 2005-08-29
- 性别: GG
- 专业: 碳素材料与超硬材料
★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★
ghcacj(金币+20):谢谢 2010-12-04 17:09:24
ghcacj(金币+20):谢谢 2010-12-04 17:09:24

2楼2010-12-04 15:07:09
sg18408926
至尊木虫 (著名写手)
- 应助: 0 (幼儿园)
- 金币: 10945.9
- 散金: 246
- 红花: 1
- 帖子: 1075
- 在线: 431.4小时
- 虫号: 727956
- 注册: 2009-03-21
- 性别: GG
- 专业: 凝聚态物性 II :电子结构
3楼2010-12-04 17:27:08
qphll
金虫 (正式写手)
- 模拟EPI: 10
- 应助: 18 (小学生)
- 金币: 2708.7
- 散金: 2294
- 红花: 8
- 帖子: 554
- 在线: 227.1小时
- 虫号: 89654
- 注册: 2005-08-29
- 性别: GG
- 专业: 碳素材料与超硬材料
★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★
ghcacj(金币+20):谢谢 2010-12-10 10:40:58
ghcacj(金币+20):谢谢 2010-12-10 10:40:58

4楼2010-12-07 23:19:38
qphll
金虫 (正式写手)
- 模拟EPI: 10
- 应助: 18 (小学生)
- 金币: 2708.7
- 散金: 2294
- 红花: 8
- 帖子: 554
- 在线: 227.1小时
- 虫号: 89654
- 注册: 2005-08-29
- 性别: GG
- 专业: 碳素材料与超硬材料
★ ★ ★ ★ ★ ★ ★ ★ ★ ★
ghcacj(金币+10, 模拟EPI+1):谢谢 2010-12-10 10:41:17
ghcacj(金币+10, 模拟EPI+1):谢谢 2010-12-10 10:41:17
|
更新一下, 稍微修改了一下前楼的程序. 现在如果在计算中有发生过反应, 那么分析程序会将那部分信息输出到另外的一个文件中. !# DEC.9, 2010 !# QPHLL !# www.muchong.com !# !# This is a program to read the output from 'fix reax/bond', TPRD, Lammps !# The output is saved into the file "bonds.connect", where each image is divided !# into three parts: !# !# (1) Head, 7 Lines; !# (2) Body, No._of_atom Lines; !# (3) Tail, 1 Line !# !# The total number of images is related with the output frequence and number of iterations. !# In this case, it is "number of iteration+1". !# !# Each line in Body part is made up of the following parameters: !# id, type, nb, id_1, id_2, ... id_nb, mol, bo_1, bo_2, ... bo_nb, abo, nlp, q !# abo = atomic bond order !# nlp = number of lone pairs !# q = atomic charge !# !# PLEASE DOUBLE CHECK YOUR OWN LAMMPS INPUT SCRIPT & OUTPUT AND MAKE CORRESPONDING CHSNGES program main implicit none integer I, J, K, L integer image, natom integer headline, tailline integer id, atype, nb, bd1, bd2, bd3, bd4, mol double precision bo1, bo2, bo3, bo4, abo, nlp, q open (unit=10, file='bonds.connect') open (unit=20, file='N129.txt', status='unknown') open (unit=21, file='N133.txt', status='unknown') open (unit=22, file='N137.txt', status='unknown') open (unit=23, file='N141.txt', status='unknown') open (unit=24, file='N145.txt', status='unknown') open (unit=25, file='N149.txt', status='unknown') open (unit=26, file='N153.txt', status='unknown') open (unit=27, file='N157.txt', status='unknown') open (unit=30, file='reactionRecord.txt', status='unknown') !# Make changes accordingly. image = 2000 headline = 7 tailline = 1 natom = 160 do I = 1, image+1 ! Skip the head part do J = 1, headline read(10,*) end do ! Each image has 'natom' lines do K = 1, natom ! read in the first three number each line to determine: ! (1) what type of atom it is, atype ! the correspondence in Lammps: 1-C, 2-H, 3-O, 4-N, 5-S ! (2) how many bonds it has, nb ! this 'nb' determines the following bond_link information & bond_order paramaters of the same line read(10,*) id, atype, nb ! TEST ! write(*,*) id, atype, nb if (atype .eq. 4) then backspace 10 ! Should have some easier way to replace this "IF", I am just toooo lazy. ! Thanks to the fact that the maximum number of bonds is 4. ^-^ !??? is it possible that nb = 0 ??? KEEP THAT IN MIND. if (nb.eq.0) then read(10,*) id, atype, nb, mol, abo, nlp, q if (id .eq. 129) then write(20, 200) id, atype, nb, mol, abo, nlp, q elseif (id .eq. 133) then write(21, 200) id, atype, nb, mol, abo, nlp, q elseif (id .eq. 137) then write(22, 200) id, atype, nb, mol, abo, nlp, q elseif (id .eq. 141) then write(23, 200) id, atype, nb, mol, abo, nlp, q elseif (id .eq. 145) then write(24, 200) id, atype, nb, mol, abo, nlp, q elseif (id .eq. 149) then write(25, 200) id, atype, nb, mol, abo, nlp, q elseif (id .eq. 153) then write(26, 200) id, atype, nb, mol, abo, nlp, q elseif (id .eq. 157) then write(27, 200) id, atype, nb, mol, abo, nlp, q 200 format(4I4, 3f14.3) endif ! If bd .ne. 3, it measn reaction is happening to Nitrogen atom. write (30, 300) I, id, atype, nb, mol, abo, nlp, q 300 format(5I4, 3f14.3) elseif (nb.eq.1) then read(10,*) id, atype, nb, bd1, mol, bo1, abo, nlp, q if (id .eq. 129) then write(20, 201) id, atype, nb, bd1, mol, bo1, abo, nlp, q elseif (id .eq. 133) then write(21, 201) id, atype, nb, bd1, mol, bo1, abo, nlp, q elseif (id .eq. 137) then write(22, 201) id, atype, nb, bd1, mol, bo1, abo, nlp, q elseif (id .eq. 141) then write(23, 201) id, atype, nb, bd1, mol, bo1, abo, nlp, q elseif (id .eq. 145) then write(24, 201) id, atype, nb, bd1, mol, bo1, abo, nlp, q elseif (id .eq. 149) then write(25, 201) id, atype, nb, bd1, mol, bo1, abo, nlp, q elseif (id .eq. 153) then write(26, 201) id, atype, nb, bd1, mol, bo1, abo, nlp, q elseif (id .eq. 157) then write(27, 201) id, atype, nb, bd1, mol, bo1, abo, nlp, q 201 format(5I4, 4f14.3) endif ! If bd .ne. 3, it measn reaction is happening to Nitrogen atom. write (30, 301) I, id, atype, nb, bd1, mol, bo1, abo, nlp, q 301 format(6I4, 4f14.3) elseif (nb.eq.2) then read(10,*) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q if (id .eq. 129) then write(20, 202) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q elseif (id .eq. 133) then write(21, 202) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q elseif (id .eq. 137) then write(22, 202) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q elseif (id .eq. 141) then write(23, 202) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q elseif (id .eq. 145) then write(24, 202) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q elseif (id .eq. 149) then write(25, 202) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q elseif (id .eq. 153) then write(26, 202) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q elseif (id .eq. 157) then write(27, 202) id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q 202 format(6I4, 5f14.3) endif ! If bd .ne. 3, it measn reaction is happening to Nitrogen atom. write (30, 302) I, id, atype, nb, bd1, bd2, mol, bo1, bo2, abo, nlp, q 302 format(7I4, 5f14.3) elseif (nb.eq.3) then read(10,*) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q if (id .eq. 129) then write(20, 203) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 133) then write(21, 203) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 137) then write(22, 203) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 141) then write(23, 203) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 145) then write(24, 203) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 149) then write(25, 203) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q elseif (id .eq. 153) then write(26, 203) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bd3, abo, nlp, q elseif (id .eq. 157) then write(27, 203) id, atype, nb, bd1, bd2, bd3, mol, bo1, bo2, bo3, abo, nlp, q 203 format(7I4, 6f14.3) endif elseif (nb.eq.4) then read(10,*) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q if (id .eq. 129) then write(20, 204) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q elseif (id .eq. 133) then write(21, 204) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q elseif (id .eq. 137) then write(22, 204) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q elseif (id .eq. 141) then write(23, 204) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q elseif (id .eq. 145) then write(24, 204) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q elseif (id .eq. 149) then write(25, 204) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q elseif (id .eq. 153) then write(26, 204) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bd3, bo4, abo, nlp, q elseif (id .eq. 157) then write(27, 204) id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q 204 format(8I4, 7f14.3) endif ! If bd .ne. 3, it measn reaction is happening to Nitrogen atom. write (30, 304) I, id, atype, nb, bd1, bd2, bd3, bd4, mol, bo1, bo2, bo3, bo4, abo, nlp, q 304 format(9I4, 7f14.3) ! Corresponding to "if (nb.eq.0) then " endif ! Corresponding to "if (atype .eq. 4) then" endif enddo do L =1,tailline read(10,*) enddo enddo end program main |

6楼2010-12-10 03:29:50
qphll
金虫 (正式写手)
- 模拟EPI: 10
- 应助: 18 (小学生)
- 金币: 2708.7
- 散金: 2294
- 红花: 8
- 帖子: 554
- 在线: 227.1小时
- 虫号: 89654
- 注册: 2005-08-29
- 性别: GG
- 专业: 碳素材料与超硬材料

7楼2010-12-11 10:32:02
hbcsucy
木虫 (小有名气)
AAA
- 应助: 0 (幼儿园)
- 金币: 5593.2
- 散金: 10
- 红花: 1
- 帖子: 293
- 在线: 192.4小时
- 虫号: 1067102
- 注册: 2010-07-31
- 性别: GG
- 专业: 凝聚态物性 II :电子结构

8楼2010-12-13 16:11:36
qphll
金虫 (正式写手)
- 模拟EPI: 10
- 应助: 18 (小学生)
- 金币: 2708.7
- 散金: 2294
- 红花: 8
- 帖子: 554
- 在线: 227.1小时
- 虫号: 89654
- 注册: 2005-08-29
- 性别: GG
- 专业: 碳素材料与超硬材料
★ ★ ★ ★ ★ ★ ★ ★ ★ ★
ghcacj(金币+10, 模拟EPI+1): 谢谢 2011-02-24 13:30:03
ghcacj(金币+10, 模拟EPI+1): 谢谢 2011-02-24 13:30:03
|
好久没有做什么更新, 这两个月来在Lammps, 尤其是reaxff上面花了不少时间, 写了不少小程序, 现在基本到一个尾声. 很多程序写的时候一鼓作气, 没有很好地加入备注, 今天花了一整天的时间注释了一些程序的用途. 有可能会慢慢发出来. 这是才写下的一段话, 是对于lammps+reax中最重要两个输出文件的处理意见. 程序本身对别人没有什么帮助, 但是这段备注我想和大家分享. ! The "REAXFRAG" code needs two files: ! (1) coordinate table <---- "molfra.configure" <----- from Lammps "molfra" dump file, prepared by this code ! (2) bond order table <---- "bonds.connect" <---- directly from Lammps 'fix' dump ! ! The corresponding commamds in "in.reaxff" file look like: ! ! "dump 1 all custom 10 molfra id type q x y z vx vy vz mass" ! "dump_modify 1 sort id" <---- This "sort" option is very important. ! ! "fix 2 all reax/bonds 10 bonds.connect" ! ! Some further explanations to the "molfra" and "bonds.connect" files. ! ! Each frame of "molfra" is made up of the head part, 9 line and body part, N line (N=number of atoms of the system). ! So an output of "m" MD iterations will produce (m+1)*(N+9) lines in "molfra". ! ! Each frame of "bonds.connect" file has head part, 7 line, body part, N line (N=number of atoms of the system) ! and tail part, 1 line, which contains a "#" sign as the ending remark. ! So an output of "m" MD iterations will produce (m+1)*(N+8) lines in "molfra". ! ! "m+1" comes from the fact that Lammps dose output the t=0 image. ! !!!!!!!!!!!! ! IT IS VERY IMPORTANT that you use same output frequency for all the dumps in Lammps. ! You also need to 'sort' the output so that atoms in different dump files are in good match. ! That explains why I prefer to seprate the minimization part from MD runs. !!!!!!!!!!!! ! ! The other input is the number of frames in 'molfra', which is the number of iterations ! you request in "in.reaxff" file, such as the following: ! ! "run 1200000" ! ! ! Required input: ! "molfra" from Lammps & number of frames in the molfra file ! ! Please always double check to make sure that is what you want to have. ! @ QPHLL, Feb.23,2010 |

9楼2011-02-24 07:13:34
qphll
金虫 (正式写手)
- 模拟EPI: 10
- 应助: 18 (小学生)
- 金币: 2708.7
- 散金: 2294
- 红花: 8
- 帖子: 554
- 在线: 227.1小时
- 虫号: 89654
- 注册: 2005-08-29
- 性别: GG
- 专业: 碳素材料与超硬材料
★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★ ★
ghcacj(金币+20): 谢谢 2011-02-24 13:30:14
ghcacj(金币+20): 谢谢 2011-02-24 13:30:14
|
这是另外一个script, 就是用来格式转化. 没有什么, 但是往往需要很多这样的script在各个软件之间作为桥梁. 有谁需要, 尽管拿去吧. ! This is to write the output XYZ file from Lammps output format into: ! (1) the normal XYZ format which is readable by most progrms, such as imol ! (2) the "initial.dat" file which is the required input when you use "read_data" ! in the "in.reaxff" file ! ! You can use this script to 'look' at each image along the way of your simulation ! or convert at any point of Lammps XYZ file into a new "initial.dat" file. ! I find it useful since I seperate minimization from MD production, and I use this ! script if there is a need for rerun a job. ! ! Be aware of the two facts: ! (1) the IDs of atoms are defined in "initial.dat" file of Lammps ! You have to compare the definition of atom with "in.reaxff" file to make sure ! that you've got the right atom types; ! ! (2) the box size is hard-coded here as: ! ! lx = 39.352194 ! ly = 42.600000 ! lz = 60.000000 ! halflz = lz/2.0 ! ! Pay attention to the PBC issue and wrap your coordinates back in one smooth image. ! ! You need to change accordingly or treat it as an interactive input. ! ! I have a different version(lmp2lmp_ver.f90), where basically I match-up ! the atom types between "initial.dat" and "in.reaxff". ! The other version(batchlmp2lmp.f90) is capable of batch conversion: ! (1) generated an index of files that you want to convert ! This can be done by this command at your target folder ! # find *.xyz > list ! (2) the format of that index file is as the following: ! Line one: Number of files (N) ! Line two to N: the name of file ! ! Required input: name of XYZ file and number of atom type ! ! Please always double check to make sure that is what you want to have. ! @ QPHLL, Feb.23,2010 ! program lmp2lmp implicit none integer natom, ntype,I, id integer zero, one,two,three,four,five double precision x, y, z,lx,ly,lz, halflz character*1 C, H, O, N, S, atype*2, ctmp1*2, ctmp2*2,ctmp3*2 character*100 filename, temp, temp2 C='C' H='H' O='O' N='N' S='S' zero=0 one=1 two=2 three=3 four=4 five=5 lx = 39.352194 ly = 42.600000 lz = 60.000000 halflz = lz/2.0 write (6,*)'Output from LAMMPS MINIMIZATION (probably structure.*.xyz) ?' read (5,'(a40)')filename temp='initial.dat'//filename temp2 ='formatted'//filename open(unit=30,file=filename,status='old') open(unit=20, file=temp, status='unknown') open(unit=40, file=temp2, status='unknown') read(30,*) natom read(30,*) write (6,*)'How many types of atoms ?' read (5,*) ntype write(40,*) natom write(40,*) write(20,*) write(20,*) write(20,*) natom, "atoms" write(20,*) write(20,*) ntype,"atom types" write(20,*) write(20,*) 0.00000,lx, "xlo xhi" write(20,*) 0.00000,ly, "ylo yhi" write(20,*) 0.00000,lz, "zlo zhi" write(20,*) write(20,*) write(20,*) "Atoms" write(20,*) do I = 1, natom read(30,*)id, x, y, z if(z.lt.halflz) then write(20,100)I,id,zero,x, y, z else write(20,100)I,id,zero,x, y, z-lz endif if(z.lt.halflz) then if (id.eq.1) then write(40,*)C, x, y, z elseif(id.eq.2) then write(40,*)H, x, y, z elseif(id.eq.3) then write(40,*)O, x, y, z elseif(id.eq.4) then write(40,*)N, x, y, z elseif(id.eq.5) then write(40,*)S, x, y, z else write(6,*)'Wrong atom ID!' stop endif else if (id.eq.1) then write(40,*)C, x, y, z-lz elseif(id.eq.2) then write(40,*)H, x, y, z-lz elseif(id.eq.3) then write(40,*)O, x, y, z-lz elseif(id.eq.4) then write(40,*)N, x, y, z-lz elseif(id.eq.5) then write(40,*)S, x, y, z-lz else write(6,*)'Wrong atom ID!' stop endif endif 100 format(I6,1x,I3,1x,I3,1x,3f12.6) enddo end program lmp2lmp |

10楼2011-02-24 07:16:55














 回复此楼
回复此楼 fly771125
fly771125