| 查看: 20374 | 回复: 68 | |||
[交流]
无意间发现两篇论文中许多句子十分相似,会是巧合吗 已有55人参与
|
|||
|
这两天阅读文献时,无意间发现两篇论文中许多句子十分相似,有些几乎一模一样,这会是巧合吗?现将两篇论文中相似句子整理如下。字母A代表论文Genome-Wide Analysis of the Dof Transcription Factor Gene Family Reveals Soybean-Specific Duplicable and Functional Characteristics,于2013年发表在PLOS ONE。字母B代表论文Comprehensive Analysis of NAC Domain Transcription Factor Gene Family in Populus trichocarpa,于2010年发表在BMC Plant Biology。 Introduction/Background (3 places) 1. A Therefore, the identification and functional characterization of TFs is essential for the reconstruction of transcriptional regulatory networks [3].In plants, ~60 families of TFs have been identified based on bioinformatics analysis and manual inspection [4,5]. The Arabidopsis genome codes for at least 1533 TFs, which account for about 5.9% of its estimated total number of genes[1]. As for soybean (Glycine max), ~12.2% of the 46,430 predicted protein-coding loci have been identified to encode 5,671 putative TFs [6]. B Thus, the identification and functional characterization of TFs is essential for the reconstruction of transcriptional regulatory networks[3]. In plants, totally more than 50 different families of TFs have been identified based on bioinformatics analysis so far [1,3]. The Arabidopsis genome encodes at least 1550 TFs, accounting for about 6.2% of its estimated total number of gene [4]. As for Populus,about6.4%of its genome was found to encode more than 2900 TFs [3,5]. 2. A Although quite a few Dof TFs have been functionally characterized in the model plant Arabidopsis and others, the functions of most members of the Dof family remain unknown. Especially in soybean, the typical legume species, there are only very limited reports on the functional characterization of Dof TFs. B Although quite a few NAC TFs have been functionally characterized in model plantsArabidopsisand rice, the functions of majority of NAC members remain unknown. Especially inPopulus, the typical model tree species, there are only very limited reports on the functional characterization of NAC TFs. 3. A In the present study, a genome-wide identification of Dof domain TFs in soybean was performed and revealed an expanded Dof family with 78 members. Detailed analysis of the sequence phylogeny, genome organization, gene structure, conserved motifs, duplication status, expression profiling, and cis-elements was performed. It is noteworthy that B In the present study, we further performed a genome-wide identification of NAC domain TFs in Populus and revealed an expanded NAC family with totally 163 members. Detailed analysis including sequence phylogeny, genome organization, gene structure, conserved motifs and expression profiling was performed. It is noteworthy that Materials and Methods (4 places) 1. A All non-redundant hits with expected values <1E-5 were collected and compared with the Dof family in PlantTFDB (http://planttfdb.cbi.edu.cn/) [5] and LegumeTFDB (http://legumetfdb.psc.riken.jp/) [36]. As for the incorrectly-predicted genes, manual re-annotation was performed using the on-line web server GENSCAN (http://genes.mit.edu/GENSCAN.html) [37] and/or RT-PCR cloning. The re-annotated sequences were further manually analyzed to confirm the presence of the Dof domain using the InterProScan program (http://www.ebi.ac.uk/Tools/InterProScan/) [38]. B All non-redundant hits with expected values less than 1.0 were collected and then compared with the NAC family in PlnTFDB http://plntfdb.bio.uni-potsdam.de/v3.0/[3] and DPTF http://dptf.cbi.pku.edu.cn/[58]. As for the incorrectly predicted genes, manual reannotation was performed using online web server FGENESH http://linux1.softberry.com/berry.phtml[59]. The reannotated sequences were further manually analyzed to confirm the presence of NAM domain using InterProScan program http://www.ebi.ac.uk/Tools/InterProScan/[60]. 2. A Unrooted phylogenetic trees were constructed with MEGA 4.0 using the Neighbor-Joining (NJ) method and the bootstrap test carried out with 1000 iterations [41]. The pairwise gap deletion mode was used to ensure that the more divergent C-terminal domains could contribute to the topology of the NJ tree. B The unrooted phylogenetic trees were constructed with MEGA 4.0 using the NeighborJoining (NJ), Minimal Evolution (ME) and Maximum Parsimony (MP) methods and the bootstrap test carried out with 1000 iterations [91]. Pairwise gap deletion mode was used to ensure that the more divergent C-terminal domains could contribute to the topology of the NJ tree. 3. A The Gene Structure Display Server program [42] was used to illustrate the exon/intron organization for individual Dof genes by comparison of the coding sequences with their corresponding genomic DNA sequences from Phytozome(http://www.phytozome.net/gmax). B Gene structure display server (GSDS) program [92] was used to illustrate exon/intron organization for individual NAC genes by comparison of the cDNAs with their corresponding genomic DNA sequences from Phytozome http://www.phytozome.net/poplar. 4. A Structural motif annotation was performed using the SMART (http://smart.embl-heidelberg.de) [49] and Pfam (http://pfam.sanger.ac.uk) databases [50]. B Structural motif annotation was performed using the SMART http://smart.embl-heidelberg.de[95] and Pfam http://pfam.sanger.ac.uk databases [96]. Results and Discussion (15 places) 1. A The identified GmDof genes encode peptides ranging from 147 to 555 amino-acids in length with an average of 335. B The identified NAC genes in Populus encode proteins ranging from 117 to 718 amino acids (aa) in length with an average of 342 aa. 2. A The detailed information of the Dof family genes in soybean, including accession numbers and similarities to their Arabidopsis orthologs, as well as nucleotide and protein sequences, are listed in Table 1 and Additional Table S1. The Dof gene family in soybean is largest compared with the estimates for other plant species, which range from ~36 in Arabidopsis [13], ~30 in rice [8], ~28 in sorghum [11] and ~27 in Brachypodium distachyon [54]. The member of Dofgenes in soybean is roughly 2.4-fold that in Arabidopsis, which is consistent with the ratio of 1.4-1.6 putative Populus homologs for each Arabidopsis gene, based on comparative genomics studies [9]. B The detailed information of NAC family genes inPopulus, including accession numbers and similarities to their Arabidopsis orthologs as well as nucleotide and protein sequences was listed in Table 1 and Additional file 1. The NAC gene family inPopulusis by far the largest one compared to the estimates for other plant species, which range from ~105 inArabidopsis[6], ~140 in rice[7] and ~101 in soybean [8]. The member of NAC genes in Populusis roughly 1.58 fold than that of Arabidopsis, which is in consistency with the ratio of 1.4~1.6 putative Populus homologs for each Arabidopsis gene based on comparative genomics studies [5]. 3. A To examine the phylogenetic relationships among the Dof domain proteins in soybean, an unrooted tree was constructed from alignments of the full-length amino-acid sequences of all GmDof proteins (Figure 2A). B To examine the phylogenetic relationship among the NAC domain proteins inPopulus, Arabidopsisand rice, an unrooted tree was constructed from alignments of the full-length NAC protein sequences (Figure 1). 4. A It is well known that gene structural diversity is a possible mechanism for the evolution of multigene families. In order to gain further insight into the structural diversity of Dof genes, we compared the exon/intron organization in the coding sequences of individual Dof genes in soybean. A detailed illustration of the exon/intron structures is shown in Figure 2B. B It is well known that gene structural diversity is a possible mechanism for the evolution of multigene families. In order to gain further insights into the structural diversity of NAC genes, we compared the exon/intron organization in the coding sequences of individual NAC genes in Populus. A detailed illustration of the exon/intron structures was shown in Figure 3B. 5. A In contrast, the gene structure appeared to be more variable in subgroups I, V and VI, which had the largest numbers of exon/intron structural variants with striking distinctions. B In contrast, the gene structure appeared to be more variable in subfamilies II and V, which had the largest number of exon/intron structure variants with striking distinctions. 6. A Substantial clustering of Dof genes was evident on several chromosomes, especially on those with high densities of the genes. B Substantial clustering of Populus NAC genes was evident on several LGs, especially on those with high densities of NAC genes. 7 A Previous studies revealed that the soybean genome has undergone at least two rounds of genome-wide duplication followed by multiple segmental duplication, tandem duplication, and transposition events such as retroposition and replicative transposition [58]. B Previous studies revealed that Populus genome had undergone at least three rounds of genome-wide duplications followed by multiple segmental duplication, tandem duplication, and transposition events such as retroposition and replicative transposition [5,68]. 8. A The distributions of Dof genes relative to the corresponding duplicate genomic blocks are illustrated in Figure 3. Within the duplicated blocks associated with a duplication event, 22 out of 38 putative paralogous pairs were preferentially-retained duplicates that were located in a segmental duplication of a long fragment (>1 Mb), B The distributions of NAC genes relative to the corresponding duplicate genomic blocks were illustrated in Figure 2. Within the 36 identified duplicated blocks associated with the recent salicoid duplication event, about 73% (87 of 120) of Populus NACs were preferentially retained duplicates that locate in both duplicated regions of 28 block pairs, 9. A In general, the Dof members demonstrated an interspersed distribution in most subfamilies, indicating that the expansion of Dof genes occurred before the divergence of soybean, Arabidopsis, and rice. B In general, the NAC members demonstrated an interspersed distribution in majority of subfamilies, indicating that the expansion of NAC genes occurred before the divergence of Populus, Arabidopsis and rice. 10. A Inspection of the phylogenetic tree topology revealed several pairs of Dof proteins with a high degree of homology in the terminal nodes of each subgroup, suggesting that they are putative paralogous pairs (Figure 2A). A total of 38 pairs of putative paralogous Dof proteins were identified, accounting for nearly the entire family (except for GmDof17.4 and GmDof05.4), with sequence identity ranging from 72% to 97% (see Additional Table S2 for details). B Inspection of the phylogenetic tree topology revealed several pairs of NAC proteins with a high degree of homology in the terminal nodes of each subfamily, suggesting that they are putative paralogous pairs (homologous genes within a species that diverged by gene duplication) (Figure 1 and Additional file 3). Totally, forty-nine pairs of putative paralogous NAC proteins were identified, accounting for more than 60% of the entire family, with sequence identities ranging from 71% to 97% (see Additional file 5 for details). 11. A To reveal the diversification of Dof genes in soybean, putative motifs were predicted by the program MEME (Multiple Em for Motif Elicitation), and a total of 30 conserved motifs were found in all the 78 Dof proteins (Figure 5). B To further reveal the diversification of NAC genes in Populus, putative motifs were predicted by the program MEME (Multiple Expectation Maximization for Motif Elicitation), and 20 distinct motifs were identified. 12. A As expected, most of the closely-related members in the phylogenetic tree had common motif compositions. B As expected, most of the closely related members in the phylogenetic tree had common motif compositions 13. A Exon/intron structures of Dof genes from soybean. Exons are represented by green boxes and introns by black lines. The sizes of exons and introns can be estimated using the scale below. B Exon/intron structures of NAC genes from Populus. Exons and introns are represented by green boxes and black lines, respectively. The sizes of exons and introns can be estimated using the scale at bottom. 14. A The deduced full length amino-acid sequences of 78 soybean, 36 Arabidopsis and 30 rice Dof genes were aligned by Clustal X 1.83 and the phylogenetic tree was constructed using MEGA 4.0 by the neighbor-joining method with 1,000 bootstrap replicates. Each Dof subgroup is indicated by a specific color. B The deduced full-length amino acid sequences of 163Populus, 105 Arabidopsis and 140 rice NAC genes were aligned by Clustal X 1.83 and the phylogenetic tree was constructed using MEGA 4.0 by the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. Each NAC subfamily was indicated in a specific color. 15. A The expression data were gene-wise normalized and hierarchically clustered. The relative expression level of a particular gene in each row was normalized against the mean value. The color scale below represents expression values, green indicating low levels and red indicating high levels of transcript abundance. B The expression data were gene-wise normalized and hierarchical clustered based on Pearson correlation. The genes marked in the same color indicate duplicated gene pairs. The relative expression level of particular gene in each row is normalized against the maximum value. Color scale at the top of each dendrogram represents log2 expression values, green represents low level and red indicates high level of transcript abundances. Conclusion (1 place) 1. A The exon/intron structure and motif composition of the Dofs were highly conserved in each subfamily, indicating their functional conservation. The Dof genes were non-randomly distributed within and across 19 chromosomes, and a high proportion of GmDofs were preferentially-retained duplicates located on duplicated blocks. B The exon/intron structure and motif compositions of NACs were highly conserved in each subfamily, indicative of their functional conservation. The NAC genes were non-randomly distributed across 19 LGs, and a high proportion of NACs were preferentially retained duplicates located on the duplicated blocks Supporting Information/Additional material (1 place) 1. A Complete list of soybean Dof gene sequences identified in the present study.The list comprises 78 GmDof gene sequences. The amino-acid sequences were deduced from their corresponding coding sequences; the genomic DNA sequences were obtained from Phytozome. B A complete list of NAC gene sequences identified in the present study. The list comprises 163 NAC gene sequences identified. Amino acid sequences are deduced from their corresponding coding sequences and genomic DNA sequences are obtained from Phytozome http://www.phytozome.net/poplar, release 2.0. 更新: 这篇论文已经因为抄袭被撤稿了。 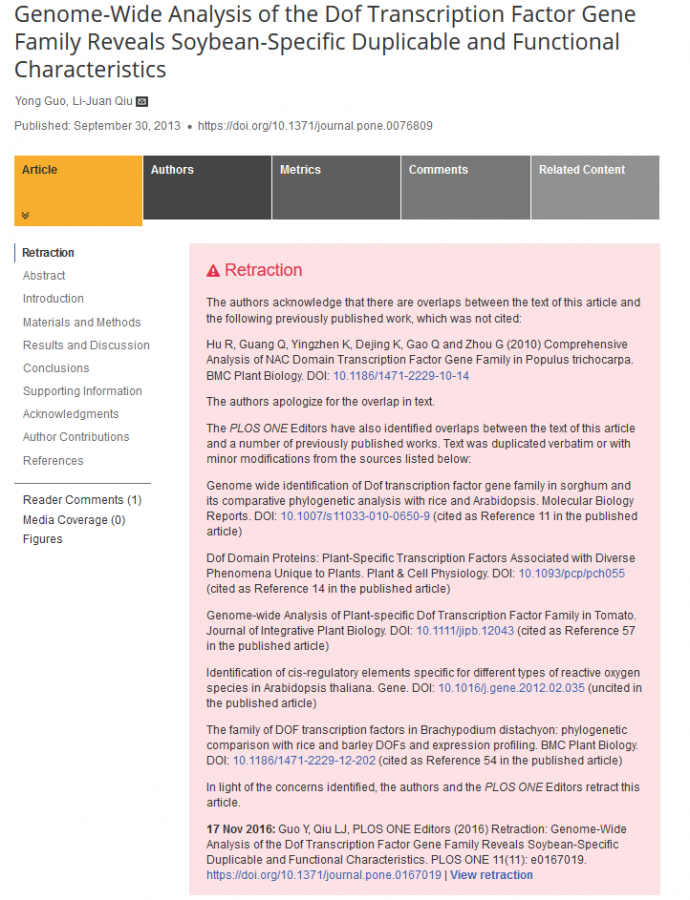 2017.png [ Last edited by waam12 on 2017-7-27 at 05:41 ] 更新:韩春雨的导师中国农科院作物所博导邱丽娟因学术抄袭被国际期刊撤稿 [ Last edited by waam12 on 2017-8-3 at 12:22 ] |
» 猜你喜欢
 网上报道青年教师午睡中猝死、熬夜猝死的越来越多,主要哪些原因引起的?
已经有3人回复
网上报道青年教师午睡中猝死、熬夜猝死的越来越多,主要哪些原因引起的?
已经有3人回复
 为什么中国大学工科教授们水了那么多所谓的顶会顶刊,但还是做不出宇树机器人?
已经有10人回复
为什么中国大学工科教授们水了那么多所谓的顶会顶刊,但还是做不出宇树机器人?
已经有10人回复
 版面费该交吗
已经有12人回复
版面费该交吗
已经有12人回复
 面上可以超过30页吧?
已经有5人回复
面上可以超过30页吧?
已经有5人回复
 体制内长辈说体制内绝大部分一辈子在底层,如同你们一样大部分普通教师忙且收入低
已经有13人回复
体制内长辈说体制内绝大部分一辈子在底层,如同你们一样大部分普通教师忙且收入低
已经有13人回复
 “人文社科而论,许多学术研究还没有达到民国时期的水平”
已经有5人回复
“人文社科而论,许多学术研究还没有达到民国时期的水平”
已经有5人回复
 什么是人一生最重要的?
已经有4人回复
什么是人一生最重要的?
已经有4人回复
» 本主题相关价值贴推荐,对您同样有帮助:
 前期工作写的论文还在审稿中,在其基础上现在的工作能不能写成论文投出去?
已经有12人回复
前期工作写的论文还在审稿中,在其基础上现在的工作能不能写成论文投出去?
已经有12人回复
 文章里的句子可以抄参考文献里的吗?
已经有21人回复
文章里的句子可以抄参考文献里的吗?
已经有21人回复
 SCI投稿过程总结
已经有124人回复
SCI投稿过程总结
已经有124人回复
 两篇论文都引用同一篇参考文献会有问题吗?
已经有11人回复
两篇论文都引用同一篇参考文献会有问题吗?
已经有11人回复
 参考文献和一篇文章有部分相同,算抄袭吗?
已经有37人回复
参考文献和一篇文章有部分相同,算抄袭吗?
已经有37人回复
 投稿意见:相似度太高
已经有14人回复
投稿意见:相似度太高
已经有14人回复
 投稿后因文章内容相似度太大被拒稿,怎么办
已经有11人回复
投稿后因文章内容相似度太大被拒稿,怎么办
已经有11人回复
 同一个人写的相关课题的两篇文章可以有内容相似的地方吗
已经有18人回复
同一个人写的相关课题的两篇文章可以有内容相似的地方吗
已经有18人回复
 关于论文内容重复及引用的问题,向大家求教!
已经有21人回复
关于论文内容重复及引用的问题,向大家求教!
已经有21人回复
 两篇文章中可以出现相同的数据吗?
已经有11人回复
两篇文章中可以出现相同的数据吗?
已经有11人回复
 自己写的两篇文章有些内容相同可以投稿吗?
已经有13人回复
自己写的两篇文章有些内容相同可以投稿吗?
已经有13人回复
 自己的后续文章引用自己的在审的文章中的句子或公式,需要标明引用吗?
已经有10人回复
自己的后续文章引用自己的在审的文章中的句子或公式,需要标明引用吗?
已经有10人回复
 SCI论文从入门到精通(第一版)
已经有93人回复
SCI论文从入门到精通(第一版)
已经有93人回复
20楼2014-10-07 06:06:46
22楼2014-10-07 06:11:09
63楼2017-07-27 07:45:26
66楼2017-07-27 12:40:13
yishuipang
铁杆木虫 (职业作家)
- 应助: 47 (小学生)
- 金币: 5720.3
- 散金: 273
- 红花: 23
- 帖子: 4549
- 在线: 486.1小时
- 虫号: 458628
- 注册: 2007-11-13
- 专业: 信息处理方法与技术

8楼2014-10-06 18:16:09
goddiao
金虫 (小有名气)
- 应助: 65 (初中生)
- 金币: 2291.5
- 散金: 100
- 红花: 14
- 帖子: 195
- 在线: 374.2小时
- 虫号: 2975938
- 注册: 2014-02-18
- 性别: GG
- 专业: 模式识别

4楼2014-10-06 16:53:12
34楼2014-10-07 15:37:05
35楼2014-10-07 17:30:49
makunjida
至尊木虫 (知名作家)
- 应助: 1291 (讲师)
- 金币: 18089.3
- 散金: 338
- 红花: 38
- 帖子: 7545
- 在线: 399小时
- 虫号: 1578528
- 注册: 2012-01-15
- 专业: 计算机软件
2楼2014-10-06 15:19:45
14楼2014-10-06 20:04:26
chenenslogan
金虫 (正式写手)
- 应助: 201 (大学生)
- 金币: 1246.9
- 散金: 148
- 红花: 14
- 帖子: 631
- 在线: 123.4小时
- 虫号: 2664352
- 注册: 2013-09-18
- 性别: GG
- 专业: 植物遗传学

7楼2014-10-06 18:01:50
在比喻日年
至尊木虫 (职业作家)
寸理莱德
- 应助: 116 (高中生)
- 金币: 20807.9
- 散金: 146
- 红花: 8
- 帖子: 3480
- 在线: 231.8小时
- 虫号: 2662169
- 注册: 2013-09-17
- 性别: GG
- 专业: 高分子材料结构与性能

36楼2014-10-07 18:57:56
tiangauyue
铁杆木虫 (著名写手)
- 应助: 38 (小学生)
- 金币: 11269.5
- 散金: 216
- 红花: 4
- 帖子: 1508
- 在线: 164小时
- 虫号: 2884712
- 注册: 2013-12-20
- 性别: GG
- 专业: 遥感机理与方法

16楼2014-10-07 00:24:48

28楼2014-10-07 09:26:16
liuzhimou
金虫 (正式写手)
- 应助: 9 (幼儿园)
- 金币: 2767.9
- 红花: 4
- 帖子: 782
- 在线: 127.8小时
- 虫号: 426825
- 注册: 2007-07-28
- 性别: GG
- 专业: 生物材料
10楼2014-10-06 18:58:55
meryl9
金虫 (小有名气)
- 应助: 2 (幼儿园)
- 金币: 1172.2
- 红花: 1
- 帖子: 264
- 在线: 41.4小时
- 虫号: 92347
- 注册: 2005-09-09
- 性别: GG
- 专业: 半导体科学与信息器件

15楼2014-10-06 20:05:07
atogle
金虫 (著名写手)
- 应助: 4 (幼儿园)
- 金币: 637.7
- 帖子: 1359
- 在线: 228.2小时
- 虫号: 232729
- 注册: 2006-03-28
- 性别: GG
- 专业: 生物化工与食品化工
19楼2014-10-07 02:57:31
rainbow3311
铁虫 (小有名气)
- 应助: 15 (小学生)
- 金币: 136.1
- 散金: 20
- 红花: 2
- 帖子: 261
- 在线: 62.3小时
- 虫号: 2164622
- 注册: 2012-12-04
- 专业: 生物电子学与生物信息处理
26楼2014-10-07 08:40:30
33楼2014-10-07 14:48:54

37楼2014-10-07 20:22:17
wgw1987come
木虫 (正式写手)
- 应助: 0 (幼儿园)
- 金币: 2030.5
- 散金: 19
- 红花: 2
- 帖子: 594
- 在线: 151.6小时
- 虫号: 933853
- 注册: 2009-12-27
- 专业: 天然药物化学
41楼2014-10-08 08:42:00
shinbade
铁杆木虫 (小有名气)
- 应助: 0 (幼儿园)
- 金币: 9061.6
- 帖子: 216
- 在线: 88.6小时
- 虫号: 93068
- 注册: 2005-09-12
- 性别: GG
- 专业: 化工系统工程
68楼2018-09-04 22:10:10
nono2009
超级版主 (文学泰斗)
No gains, no pains.
-

专家经验: +21105 - SEPI: 10
- 应助: 28684 (院士)
- 贵宾: 513.911
- 金币: 2555220
- 散金: 27828
- 红花: 2147
- 沙发: 66666
- 帖子: 1602255
- 在线: 65200.9小时
- 虫号: 827383
- 注册: 2009-08-13
- 性别: GG
- 专业: 工程热物理与能源利用
- 管辖: 科研家筹备委员会
3楼2014-10-06 16:21:56
绍敏郡主
木虫 (著名写手)
- 应助: 289 (大学生)
- 金币: 1251.1
- 散金: 2196
- 红花: 41
- 帖子: 1575
- 在线: 1866.7小时
- 虫号: 2611526
- 注册: 2013-08-22
- 专业: 原子和分子物理
5楼2014-10-06 16:53:45
6楼2014-10-06 17:52:10
bifurcation
木虫 (正式写手)
- 应助: 5 (幼儿园)
- 金币: 2333.7
- 散金: 430
- 红花: 5
- 帖子: 338
- 在线: 578.2小时
- 虫号: 2149928
- 注册: 2012-11-27
- 专业: 偏微分方程
9楼2014-10-06 18:42:30














 回复此楼
回复此楼

