| 查看: 13760 | 回复: 66 | |||||||||||||||||
daxu木虫 (正式写手)
|
[交流]
【分享】fullprof精修的简单步骤和例子--——供初学者参考已有36人参与
|
||||||||||||||||
|
Fullprof Refinement - Y2O3 Files needed: y2o3.dat; y2o3.cif Learning Outcomes: This example shows a simple Rietveld refinement of Y2O3 in fullprof. You may have used the same dataset in topas/gsas. 1. Create a new folder and copy the data file Y2O3.dat and the Y2O3.cif files into it. Start WinPlotr and press the EdPCR button. Press cif-pcr, select Y2O3.cif and press ok in the window that appears. The next step is to work down each of the buttons. 2. Press General and type in a title. 3. Press Patterns, Data file/peak shape and select the data file Y2O3.dat. Select X,Y,sigma (XYdata) as data format. 3.1 In Refinement/simulation select X-ray; make sure, the correct radiation/wavelength is chosen. 3.2 Pattern calculation/ peak shape: Select e.g. Pseudo-Voigt (scroll to find this); change the Range of calculation to a reasonable cut-off value. Higher values will result in longer calculation times, too low values will lead to poor fits. Try with a value of e.g. 15*FWHM. Compare the result to that using a value of 2*FWHM. 4. Select the Background Type, chose e.g. 6-coefficient polynomial. 5. You can use Excluded Regions to exclude selected 2theta ranges from the refinement. 6. Return to the main EdPCR window and click on Phases. Type in the phase name, e.g. Y2O3. Select the Method of calculation, e.g. Structural Model (Rietveld Method) for Rietveld, Profile matching with constant scale factor for LeBail fits. 7. Press Contribution to patterns and select X-ray. Select Pseudo-Voigt Peak shape. 8. Press Refinement in the main window. Chose an adequate number of Refinement cycles. Start with e.g. 10 cycles. Tick the parameters you want to refine in the different sections of this Refinement menu. Select a reasonable order for refining the parameters and make sure to gradually increase the number of parameters. Selecting too many parameters in one step will lead to divergence of the refinement. Start this refinement by selecting the lattice parameters, profile parameter W, Scale factor (from the "Profile" box) and the first two background polynomial coefficients (in background box). If you are happy with your selection press OK and save the input file (file/save) u into the same folder as the data file. There is no need to close the EdPCR window. N.B. if you don't click save the pcr file will not be updated. 9. It's might be worth going through the pcr file to check all your changes have been made at this stage. In particular check that all three atoms in the cif file (2*Y and 1*O) have been loaded. 10. Press FP in Winplotr, select input and data file. The refinement will start and inform you about the residuals, chi^2 and the parameter with the largest shift in each cycle. Continue with this data set until convergence is reached. If the refinement diverges, think of a different refinement strategy. Click on "OK" while you want to continue refining and "No" when you're happy. 11. You can look at the refinement by zooming in the winplotr window. A right click returns you to an unzoomed picture. 12. After convergence, reload the input file in EdPCR (File/reload). N.B. if you don't do this you won't get the refined values. 13. Visually inspect the refinement result and think of a strategy to improve the fit. Select additional parameters to refine, e.g. U and V and/or more background coefficients. Save the input file and redo the refinement… Continue until you are satisfied with the refinement result. Regularly check the parameter values to identify fit artefacts quickly. If the parameters have physically meaningless values, think of another refinement strategy. Pretty often, a different order of refining the parameters does the trick. Make sure to reset the parameters to reasonable values before trying out a different strategy. 14. Prior to refining asymmetry parameters, open the pcr-file by clicking on the PCR-button in Winplotr. Manually enter a reasonable cut-off 2 theta value for asymmetry-correction below the flag AsyLim (try different values and inspect the refinement results). Save the pcr and return to the EdPCR window. Reload the input file and sequentially start to refine asymmetry parameters. Make sure that your selection of the peak-profile function is compatible with your selection of asymmetry correction. Some profile functions already have an implemented asymmetry correction. 15. Play with the output options to check, what information is obtained. If you want to do some Fourier-map calculation to e.g., find missing atoms, select Patterns output - Structure factors file (FOU) and tick GFOURIER. You can then use the program GFOURIER to calculate different Fourier-maps. 16. If you exclude the 2 theta region between 0 and 16°, you should finally obtain an Rwp of around 6% and a Bragg-R factor for Y2O3 around 3.5%. Modified 18-Jun-2009 by John S.O. Evans. 另外,我再把这个网址http://www.dur.ac.uk/john.evans/ ... torial_fullprof.htm 和我精修的结果附上,供大家讨论交流,共同学习~~~ 关于y2o3.dat; y2o3.cif 这两个文件,大家可以在我提供的网址里下载,精修结果图见链接http://g.zhubajie.com/urllink.php?id=854077667qc4np6tbinuklm 另外精修的一般顺序为: scalor->零点矫正->晶胞参数->背底->峰型参数->温度因子->原子坐标、占位等。 |
» 收录本帖的淘贴专辑推荐
 Lithium-ion Batteries锂离子电池资料集锦 Lithium-ion Batteries锂离子电池资料集锦 |  化学电源之锂离子电池 化学电源之锂离子电池 |  锂电池材料篇 锂电池材料篇 |  精华帖-好资源 精华帖-好资源 |
 Lithium ion batterie Lithium ion batterie |  晶体结构精修 晶体结构精修 |  结构分析软件学习 结构分析软件学习 |  微结构与性能的关系 微结构与性能的关系 |
 结构精修 结构精修 |  科研 科研 |  PXRD相关 PXRD相关 |  专用帖子集 专用帖子集 |
 分析测试 分析测试 |  son of bitch 的精修 son of bitch 的精修 |
» 本帖已获得的红花(最新10朵)
» 猜你喜欢
 求南京大学《物理化学》第六版上下册和对应物理化学学习辅导
已经有0人回复
求南京大学《物理化学》第六版上下册和对应物理化学学习辅导
已经有0人回复
 江苏理工学院资源与环境学院 资源与环境专硕接收2024年级调剂生
已经有1人回复
江苏理工学院资源与环境学院 资源与环境专硕接收2024年级调剂生
已经有1人回复
 无机化学论文润色/翻译怎么收费?
已经有187人回复
无机化学论文润色/翻译怎么收费?
已经有187人回复
 求助MIL-125(Ti) 的CIF文件
已经有0人回复
求助MIL-125(Ti) 的CIF文件
已经有0人回复
 求助MIL-125(Ti)的CIF 文件
已经有1人回复
求助MIL-125(Ti)的CIF 文件
已经有1人回复
 美国 Tufts University, Yu Zhang课题组博士后招聘(金属有机/无机合成及催化)
已经有9人回复
美国 Tufts University, Yu Zhang课题组博士后招聘(金属有机/无机合成及催化)
已经有9人回复
 中原工学院邵志超副教授课题组招收调剂生2名
已经有0人回复
中原工学院邵志超副教授课题组招收调剂生2名
已经有0人回复
 我校 有两个085601 材料工程调剂名额
已经有0人回复
我校 有两个085601 材料工程调剂名额
已经有0人回复
» 本主题相关价值贴推荐,对您同样有帮助:
 用fullprof 精修的顺序是怎样的?
已经有7人回复
用fullprof 精修的顺序是怎样的?
已经有7人回复 fullprof精修 怎么计算值这么小的,内有图
已经有6人回复
fullprof精修 怎么计算值这么小的,内有图
已经有6人回复 SnO2-fullprof精修问题
已经有9人回复
SnO2-fullprof精修问题
已经有9人回复 Fullprof精修后标准差太大怎么办?
已经有3人回复
Fullprof精修后标准差太大怎么办?
已经有3人回复 fullprof怎样 用两个不同的晶系 来修同一个晶体
已经有4人回复
fullprof怎样 用两个不同的晶系 来修同一个晶体
已经有4人回复 请高手帮我用fullprof软件精修一个数据
已经有3人回复
请高手帮我用fullprof软件精修一个数据
已经有3人回复 求简单方法长针状晶体
已经有3人回复
求简单方法长针状晶体
已经有3人回复 fullprof 结构精修。有点抓狂,求帮助!!!
已经有7人回复
fullprof 结构精修。有点抓狂,求帮助!!!
已经有7人回复 求助fullprof精修后键角
已经有36人回复
求助fullprof精修后键角
已经有36人回复 Fullprof修晶胞参数
已经有5人回复
Fullprof修晶胞参数
已经有5人回复 FullProf结构精修,数据中峰有劈裂,该如何调整参数
已经有14人回复
FullProf结构精修,数据中峰有劈裂,该如何调整参数
已经有14人回复 Fullprof 精修结果Rexp很大
已经有16人回复
Fullprof 精修结果Rexp很大
已经有16人回复 50金币求fullprof中文教程
已经有11人回复
50金币求fullprof中文教程
已经有11人回复 多相粉末fullprof精修问题
已经有10人回复
多相粉末fullprof精修问题
已经有10人回复 Fullprof多相精修软件使用
已经有15人回复
Fullprof多相精修软件使用
已经有15人回复 【求助】关于Bragg peak!!
已经有6人回复
【求助】关于Bragg peak!!
已经有6人回复 【求助】PARAMETER OUT OF RANGE
已经有10人回复
【求助】PARAMETER OUT OF RANGE
已经有10人回复 【求助】fullprof 精修,指标化-欢迎参与讨论
已经有20人回复
【求助】fullprof 精修,指标化-欢迎参与讨论
已经有20人回复 【交流】关于fullprof精修结果中的几个疑问?
已经有13人回复
【交流】关于fullprof精修结果中的几个疑问?
已经有13人回复 最新Fullprof软件纳米盘下载
已经有11人回复
最新Fullprof软件纳米盘下载
已经有11人回复

daxu
木虫 (正式写手)
- 应助: 34 (小学生)
- 金币: 4419.6
- 散金: 218
- 红花: 23
- 帖子: 819
- 在线: 657.8小时
- 虫号: 574494
- 注册: 2008-06-16
- 性别: GG
- 专业: 矿物学(含矿物物理学)

6楼2010-03-23 14:25:32
daxu
木虫 (正式写手)
- 应助: 34 (小学生)
- 金币: 4419.6
- 散金: 218
- 红花: 23
- 帖子: 819
- 在线: 657.8小时
- 虫号: 574494
- 注册: 2008-06-16
- 性别: GG
- 专业: 矿物学(含矿物物理学)
daxu: 回帖置顶 2012-03-24 14:35:39
|
看到有的网友编辑pcr有问题,并且精修结果不好,我特把我修的pcr文件贴上来供大家参考,当然大家也可以进一步尝试不同的精修方式,或许得到更好的结果: COMM Y2O3 ! Current global Chi2 (Bragg contrib.) = 4.086 ! Files => DAT-file: Y2O3.dat, PCR-file: Y2O3 !Job Npr Nph Nba Nex Nsc Nor Dum Iwg Ilo Ias Res Ste Nre Cry Uni Cor Opt Aut 0 5 1 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 1 ! !Ipr Ppl Ioc Mat Pcr Ls1 Ls2 Ls3 NLI Prf Ins Rpa Sym Hkl Fou Sho Ana 0 0 1 0 1 0 4 0 0 3 10 0 0 0 0 0 0 ! ! Lambda1 Lambda2 Ratio Bkpos Wdt Cthm muR AsyLim Rpolarz 2nd-muR -> Patt# 1 1.540560 1.544390 0.50000 40.000 8.0000 0.7998 0.0000 70.00 0.0000 0.0000 ! !NCY Eps R_at R_an R_pr R_gl Thmin Step Thmax PSD Sent0 10 0.10 1.00 1.00 1.00 1.00 3.1200 0.020004 150.0000 0.000 0.000 ! ! Excluded regions (LowT HighT) for Pattern# 1 0.00 16.00 ! ! 23 !Number of refined parameters ! ! Zero Code SyCos Code SySin Code Lambda Code MORE ->Patt# 1 0.00039 21.0 0.00000 0.0 0.00000 0.0 0.000000 0.00 0 ! Background coefficients/codes for Pattern# 1 (Polynomial of 6th degree) 569.793 -50.170 182.409 -237.369 114.952 -18.432 41.00 51.00 61.00 141.00 151.00 161.00 !------------------------------------------------------------------------------- ! Data for PHASE number: 1 ==> Current R_Bragg for Pattern# 1: 3.18 !------------------------------------------------------------------------------- Y2O3 ! !Nat Dis Ang Pr1 Pr2 Pr3 Jbt Irf Isy Str Furth ATZ Nvk Npr More 3 0 0 0.0 0.0 1.0 0 0 0 0 0 3613.047 0 5 0 ! I a -3 <--Space group symbol !Atom Typ X Y Z Biso Occ In Fin N_t Spc /Codes Y1 Y 0.25000 0.25000 0.25000 0.47670 0.16667 0 0 0 0 0.00 0.00 0.00 211.00 0.00 Y2 Y 0.96753 0.00000 0.25000 0.44644 0.50000 0 0 0 0 171.00 0.00 0.00 221.00 0.00 O1 O 0.39072 0.15345 0.38024 0.19301 1.00000 0 0 0 0 181.00 191.00 201.00 231.00 0.00 !-------> Profile Parameters for Pattern # 1 ! Scale Shape1 Bov Str1 Str2 Str3 Strain-Model 0.10764E-03 0.57902 0.00000 0.00000 0.00000 0.00000 0 11.00000 131.000 0.000 0.000 0.000 0.000 ! U V W X Y GauSiz LorSiz Size-Model 0.012274 -0.013685 0.013644 0.001827 0.000000 0.000000 0.000000 0 71.000 81.000 91.000 101.000 0.000 0.000 0.000 ! a b c alpha beta gamma #Cell Info 10.602524 10.602524 10.602524 90.000000 90.000000 90.000000 31.00000 31.00000 31.00000 0.00000 0.00000 0.00000 ! Pref1 Pref2 Asy1 Asy2 Asy3 Asy4 0.00000 0.00000 0.19590 0.02896 0.00000 0.00000 0.00 0.00 111.00 121.00 0.00 0.00 ! 2Th1/TOF1 2Th2/TOF2 Pattern # 1 16.000 150.000 1 大家只需要把上面的内容复制到你的pcr文件里就可以运行。这只是一个简单的例子,精修比较容易,事实上重要的是对精修结果的判断,另外还要多看文献查资料和manual 精修之后,结果如下: ==> RELIABILITY FACTORS WITH ALL NON-EXCLUDED POINTS FOR PATTERN: 1 => Cycle: 1 => MaxCycle: 10 => N-P+C: 6678 => R-factors (not corrected for background) for Pattern: 1 => Rp: 4.35 Rwp: 5.96 Rexp: 3.11 Chi2: 3.67 L.S. refinement => Conventional Rietveld R-factors for Pattern: 1 => Rp: 9.74 Rwp: 10.3 Rexp: 5.37 Chi2: 3.67 => Deviance: 0.247E+05 Dev* : 3.699 => DW-Stat.: 0.6511 DW-exp: 1.9311 => N-sigma of the GoF: 154.392 ==> RELIABILITY FACTORS FOR POINTS WITH BRAGG CONTRIBUTIONS FOR PATTERN: 1 => N-P+C: 6001 => R-factors (not corrected for background) for Pattern: 1 => Rp: 4.28 Rwp: 5.89 Rexp: 3.04 Chi2: 3.74 L.S. refinement => Conventional Rietveld R-factors for Pattern: 1 => Rp: 9.01 Rwp: 9.86 Rexp: 5.09 Chi2: 3.74 => Deviance: 0.226E+05 Dev* : 3.768 => DW-Stat.: 0.7104 DW-exp: 1.9277 => N-sigma of the GoF: 150.354 => Global user-weigthed Chi2 (Bragg contrib.): 4.09 ----------------------------------------------------- BRAGG R-Factors and weight fractions for Pattern # 1 ----------------------------------------------------- => Phase: 1 Y2O3 => Bragg R-factor: 3.18 Vol: 1191.867( 0.007) Fract(%): 100.00( 0.40) => Rf-factor= 2.08 ATZ: 3613.047 Brindley: 1.0000 |

48楼2012-03-24 14:28:47
★ ★
小木虫(金币+0.5):给个红包,谢谢回帖交流
wsht212(金币+1):谢谢交流,希望常来! 2010-05-14 23:17:52
小木虫(金币+0.5):给个红包,谢谢回帖交流
wsht212(金币+1):谢谢交流,希望常来! 2010-05-14 23:17:52
9楼2010-05-14 20:30:59
ninezero
新虫 (小有名气)
- 应助: 1 (幼儿园)
- 金币: 65.5
- 散金: 2
- 帖子: 79
- 在线: 56.5小时
- 虫号: 712262
- 注册: 2009-03-01
- 性别: GG
- 专业: 凝聚态物性 II :电子结构
2楼2010-03-19 08:55:11
3楼2010-03-22 09:27:06
4楼2010-03-22 09:30:05
5楼2010-03-22 19:52:32
| 谢谢分享 |
7楼2010-04-01 14:04:10
myselfe123
铜虫 (正式写手)
- 应助: 7 (幼儿园)
- 金币: 173.7
- 红花: 3
- 帖子: 321
- 在线: 124.3小时
- 虫号: 758793
- 注册: 2009-04-27
- 性别: GG
- 专业: 无机非金属类光电信息与功
8楼2010-05-14 20:04:06
wsht212
荣誉版主 (文学泰斗)
江湖浪子之飞天
- CMEI: 4
- 应助: 0 (幼儿园)
- 贵宾: 5.069
- 金币: 37097.2
- 散金: 1314
- 红花: 80
- 沙发: 89
- 帖子: 476185
- 在线: 1854.3小时
- 虫号: 425501
- 注册: 2007-07-26
- 专业: 金属有机化学
- 管辖: 晶体
10楼2010-05-14 23:18:03













 回复此楼
回复此楼 yuping31216
yuping31216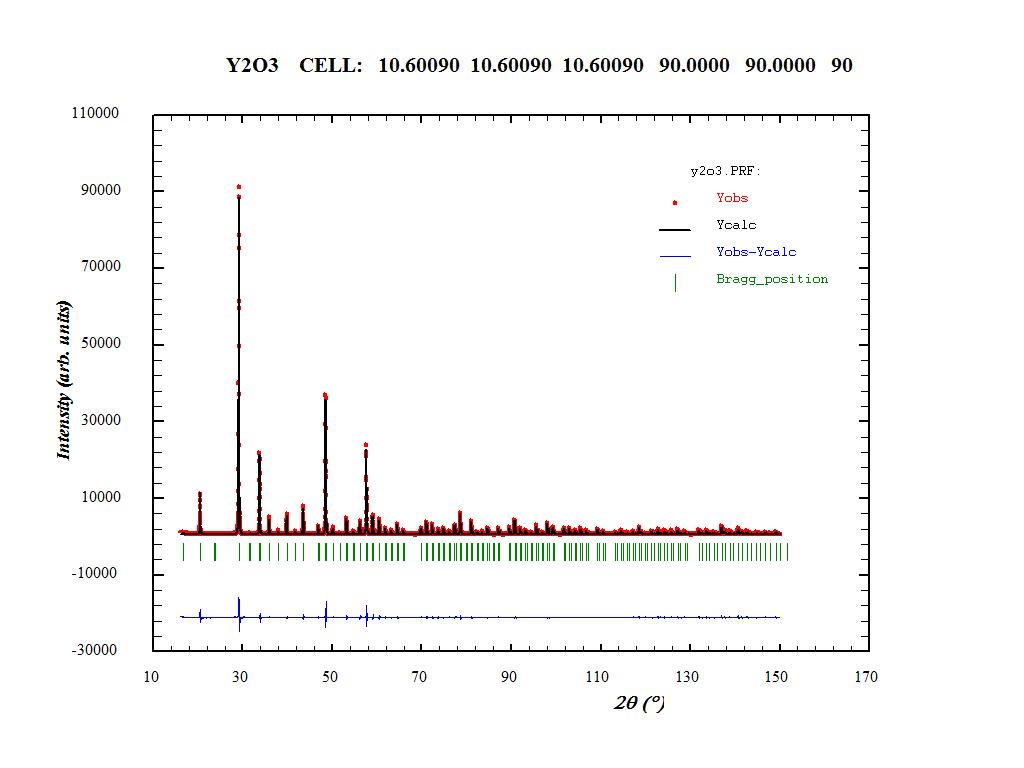






 2
2